Software

I have been developing scientific software for the most part of my career. Some of the packages I have made available here in the hope that they might be useful.
Full packages
These are relatively complex scientific applications.
MULTOVL
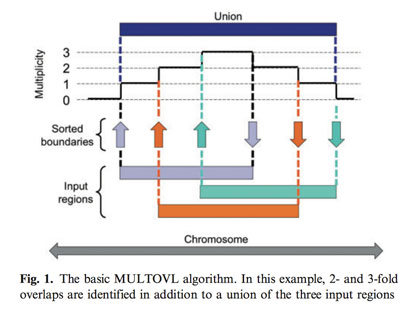
MULTOVL is a C++ application that can detect multiple overlaps of genomic regions very quickly. The basic idea of the algorithm is illustrated below.
MULTOVL User Guide
The source code (tarred-gzipped archive, 794 kB) and precompiled binaries are freely available from BitBucket and from GitHub. For HPC applications I provide a Singularity container as well. The software is referenced at OmicTools and at BioTools.
Please cite the following publication when using MULTOVL in your research:
Aszódi, A. (2012): MULTOVL: Fast multiple overlaps of genomic regions. Bioinformatics 28: 3318-3319.
SURFCOMP
This package implements a comparison and alignment of molecular surfaces based on physicochemical features mapped to surface patches. The software was written by Christian Hofbauer as part of his PhD thesis under the co-supervision of Dr Hans Lohninger (Technical University of Vienna) and myself.
Please cite the following publications when using SURFCOMP in your research:
Hofbauer, C., Lohninger, H., Aszódi, A. (2004): SURFCOMP: A novel graph-based approach to molecular surface comparison. J. Chem. Inf. Model. 44: 837-847.
Hofbauer, C., Aszódi, A. (2005): SH2 binding site comparison: A new application of the SURFCOMP method. J. Chem. Inf. Model. 45: 414-421.
OpenStructure
An object-oriented toolkit for (macro)molecular modelling, initially developed in collaboration between the Novartis In Silico Sciences (ISS) unit which I built up and led, the University of Basel and the Fachhochschule Aargau. When the In Silico Sciences unit was closed down, Novartis kindly released the source so that the other collaboration partners could continue working on it. The code is now available at the OpenStructure website and the package is described in a Bioinformatics paper. You will find my name in the Acknowledgements.
DRAGON
This is a protein structure prediction program package using distance geometry. I developed it between 1992 and 1996 when I was a postdoc in Willie Taylor's lab at the NIMR in London. I have written several papers using DRAGON, here is the main publication describing the algorithms and the software:
Aszódi, A., Gradwell, M. J. and Taylor, W. R. (1995): Global fold determination from a small number of distance restraints. J. Mol. Biol. 251, 308-326.
The DRAGON source code, written in ancient C++, is of historical interest only. It is available on GitHub.
SDPDIST
This application can reconstruct complex 3-dimensional structures from an incomplete set of distance constraints and restraints. I am still working on it. More information will be available after publication.
Utilities
Containers
- Singularity container for a toolchain to develop C++ applications with Boost. Useful for multistage builds.
- Singularity container packaging the IpOpt interior-point optimizer.
Miscellaneous tools
- WelchTest for Mathematica: The TTest function warns if the two groups to be compared have different variances, but does not run the Welch test which is appropriate for this situation. I wrote a short Mathematica function that implements the Welch test.
- cmake-python-config: A CMake snippet that can reliably detect your Python3 installation and the corresponding library locations. Use it for mixed-language programming (e.g. C++/Python projects).
- Razorback 2: This is a collection of mathematical and statistical utilities in C and C++, written mainly at Novartis. The SURFCOMP package relies on this library.
Other people's code
This is a collection of various packages written by others that I found useful. Please consult the original sources for the authors.
IpOpt easy installation package
The IpOpt nonlinear solver and its supporting libraries (HSL, Metis, Mumps) packaged together for convenience, with installation instructions.